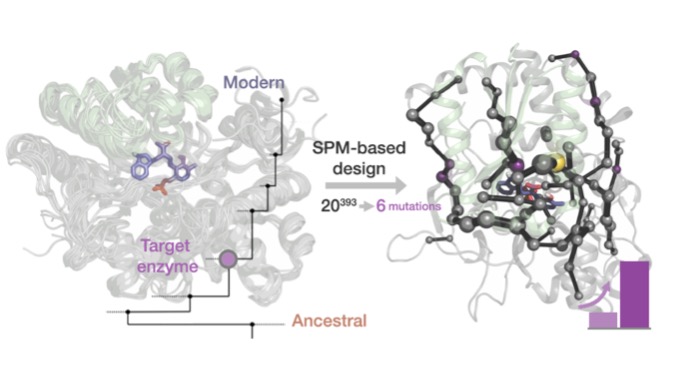
Allostery is a central mechanism for the regulation of multi-enzyme complexes. The mechanistic basis that drives allosteric regulation is poorly understood but harbors key information for enzyme engineering. In this talk, our new computational tools based on inter-residue correlations from microsecond time-scale Molecular Dynamics (MD) simulations and enhanced sampling techniques are applied in Tryptophan synthase (TrpS) complex.[1,2] TrpS is composed of TrpA and TrpB subunits, which allosterically activate each other and have no activity when isolated. [3,4] We show how distal mutations introduced in TrpS resuscitate the allosterically-driven conformational regulation and alter the populations and rates of exchange between multiple conformational states, which are essential for the multistep reaction pathway of the enzyme.[3] The exploration of the conformational landscape of TrpS is key for identifying conformationally-relevant amino acid residues of TrpB distal from the active site.[4] We predict positions crucial for shifting the inefficient conformational ensemble of the isolated TrpB to a productive ensemble through intra-subunit allosteric effects. The experimental validation of the new conformationally-driven TrpB design demonstrates its superior stand-alone activity in the absence of TrpA, comparable to those enhancements obtained after multiple rounds of experimental laboratory evolution. Our work evidences that the current challenge of distal active site prediction for enhanced function in computational enzyme design has become within reach.
Sílvia Osuna1,2*, Cristina Duran,1 Miguel Ángel Maria-Solano1, Javier Iglesias-Fernández1
1 Institut de Química Computacional i Catàlisi (IQCC) and Departament de Química, Universitat de Girona, Girona
2 ICREA, Barcelona
*silvia.osuna@udg.edu
References
[1] Maria-Solano, M. A.; Serrano-Hervás, E.; Romero-Rivera, A.; Iglesias-Fernández, J.; Osuna, S. Role of conformational dynamics for the evolution of novel enzyme function, Chem. Commun. 2018, 54, 6622-6634.
[2] Osuna, S. The challenge of predicting distal active site mutations in computational enzyme design, WIREs Comput Mol Sci. 2020, e1502.
[3] Maria-Solano, M. A.; Iglesias-Fernández, J.; Osuna, S. Deciphering the Allosterically Driven Conformational Ensemble in Tryptophan Synthase Evolution, J. Am. Chem. Soc. 2019, 141, 33, 13049-13056.
[4] Maria-Solano, M. A.; Kinateder, T.; Iglesias-Fernández, J.; Sterner, R.; Osuna, S. In Silico Identification and Experimental Validation of Distal Activity-Enhancing Mutations in Tryptophan Synthase. ACS Catal. 2021, 11, 13733-13743.
Biography

Dr. Osuna is an ICREA research professor at the Universitat de Girona (UdG). Her research is based on the computational design of enzymes. This research line is funded by a European Research Council (ERC) –Starting Grant, Spanish MINECO project, and a Human Frontier Science Program – Program Grant. She has been awarded many prestigious grants: for her post-doc at the University of California, Los Angeles (UCLA) she was awarded a Marie Curie IOF fellowship, followed by Juan de la Cierva postdoctoral fellowship, later on Ramón y Cajal position, and the 2017 ICREA position. She has also been awarded many prestigious awards: Marcial Moreno award 2021 (RSEQ-Cat), Catalan National Research Award – Young Talent 2019 from Fundació Catalana de Recerca i Innovació (FCRi), Young Investigator Award by the company Lilly and Royal Spanish Society of Chemistry RSEQ (2019), the Young Investigator Award of EuCheMS Organic Division (2017), the Young Researcher award by the Royal Spanish Society of Chemistry (RSEQ 20116), and the Research award by the Fundación Princesa de Girona (FPdGi 2016- Science category).